NMSU computer science research may help understand, analyze biological systems
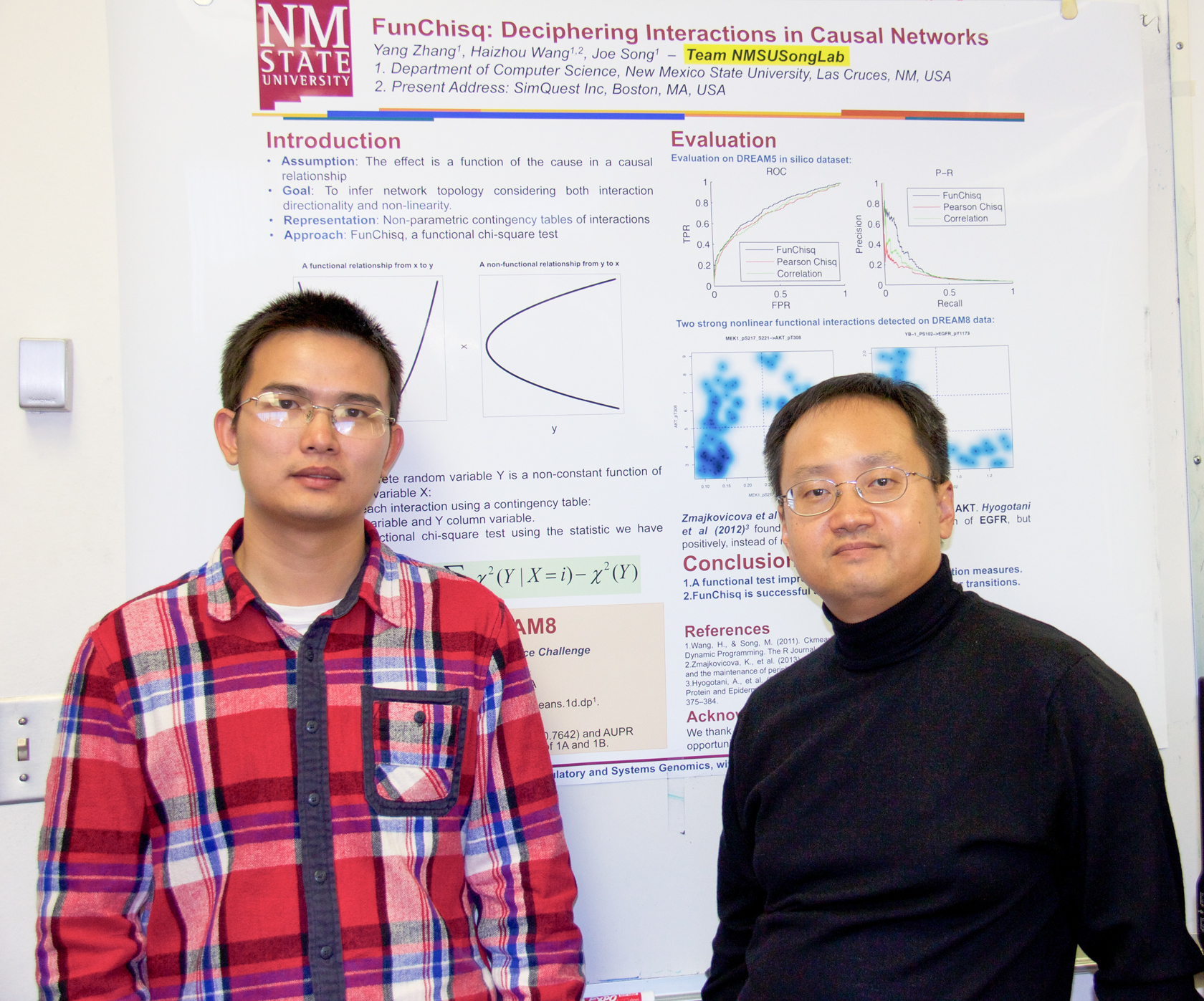
Outperforming 80 teams from around the world, Joe Song and his student, Yang Zhang, scored first place in one of three challenges in a computer science competition that encourages researchers to develop software systems that could help understand how cell reproduction leads to breast cancer.
Sponsored by Synapse, the Heritage Provider Network-Dialogue of Reverse Engineering Assessment of Methods Breast Cancer Network Inference challenges participants to create predictive algorithms in various competitions. The New Mexico State University team will present their research at the DREAM conference in Toronto Nov. 8-12.
Song, who teaches computer sciences at in NMSU's College of Arts and Sciences, explained that the goal was to develop a software system that can be broadly used for studying biological systems, although breast cancer was used as a testing platform to evaluate performance of computational methods within the challenges.
According to the Synapse website, the overall purpose of the DREAM breast cancer network inference competition, beyond the sub-challenges, "is to foster collaborations of like-minded researchers that together will find the solution for vexing problems that matter most to citizens and patients."
"Breast cancer is considered a disease contributed to by many genes," Song said. "We aim at using computers to depict how genes work together as a network in a cancer cell. Overall, our goal was to understand, at a molecular level, gene interaction; how cells duplicate and how they can cause tumor tissues to grow uncontrollably. We want to know what's leading to this cell proliferation.
"Our results have revealed which genes may cause tumors to grow unchecked, and how they may have responded to environmental stimuli. This allows biologists to gain a deeper insight to cell proliferation and to eventually identify those underlying proteins not functioning properly as drug targets."
Participants in the competition were given data generated by computer or via simulation using mathematical models of protein interactions, as well as real protein activity data from breast cancer cells. The submitted predictions were compared to factual findings to determine the most accurate predictor. The results submitted by Song and Zhang scored first place and the team will receive a $7,500 cash prize from HPN.
Other challenges involved building a network model to represent the active cell signaling pathways in breast cancer, and proposing novel strategies to visualize the high-dimensional data.
Cash incentives in this year's challenge seemed to have led to the highest number of participants yet, Song said. This was his team's fourth time taking part in the competition.
"We developed a novel functional chi-square testing approach from our methods used in previous DREAM challenges," he said. "The current method is a result of years of research efforts by our lab. Our work reduces the assumptions made in previous work."
The algorithm developed by Song and Zhang can analyze gene expression data to hunt complex interaction patterns more accurately and quickly than the human eye, they explained. Their method goes beyond correlations and aims to determine causality and changes at the protein level. The software is not restricted to biological systems, and could potentially apply to other types of networks.
"The idea was to evaluate evidence for causality innovatively," said Zhang, a doctoral student. "This challenge has real-world application potential to understand gene networks. Other approaches such as correlation do not fully explore information in the data. We look beyond correlation and other established measures of association to determine causality."
"To study complex biological systems, one often has to make assumptions because a full spectrum of activities of molecules cannot be easily observed," Song added. "Unlike a computer, which you can take apart and examine in great detail, molecular parts and connections can only be indirectly measured. Biological mechanisms, evolved over time, though far from random, are very difficult to predict."
Zhang said he was drawn to computational biology because it is a relatively new discipline that offers many opportunities for breakthroughs.
"There are many life science problems (such as treating HIV and breast cancer) that don't have a satisfactory solution yet," he said. "With the power of computers, we are trying to approach those genetic markers that may reveal where a goldmine may be located."
Zhang studied industrial design at the Beijing University of Posts and Telecommunications - Song's alma mater. The pair hopes to publish their findings soon. Their research has been previously funded by the National Science Foundation, National Cancer Institute, among other agencies.
The DREAM challenge is now in its eighth year. Past winners of the various challenges include teams from Stanford University, the Massachusetts Institute of Technology and the University of Helsinki, Finland.